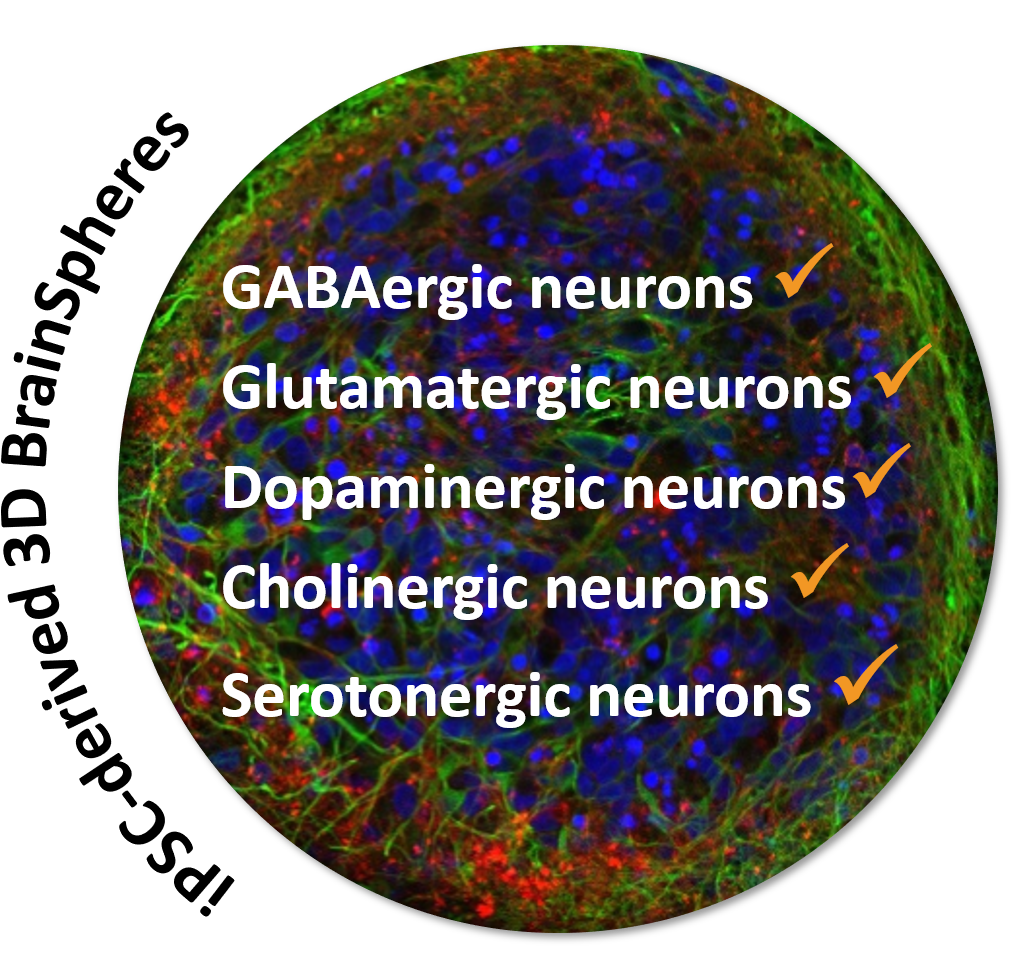
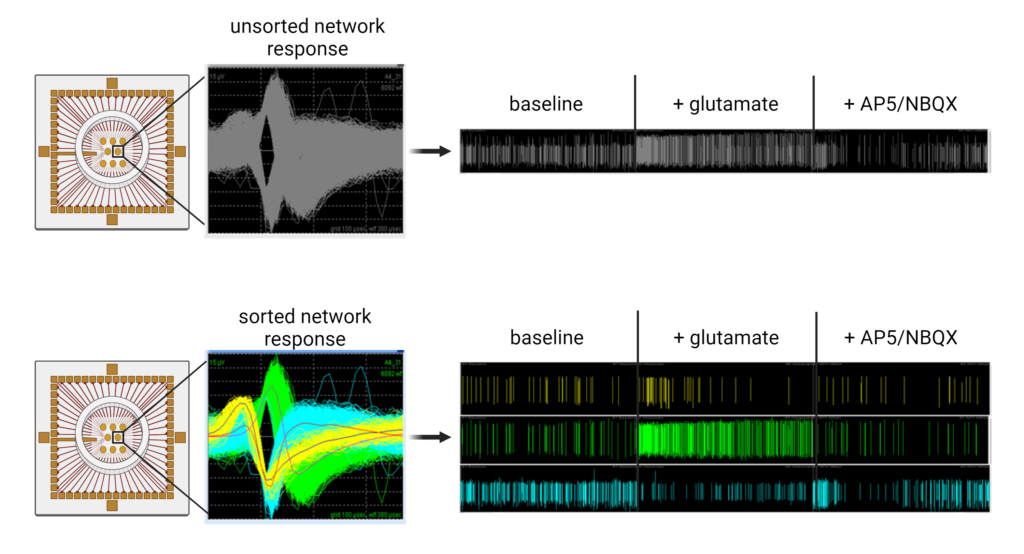
Since the nervous system is a very complex organ, it can be disrupted via a plethora of modes-of-action (MoA) involving neurotransmitter receptors and ion transporters, amongst others. In general, these MoA affect neuronal function and communication by inhibiting neurotransmitter synthesis or degradation, increasing or preventing neurotransmitter release, blocking neurotransmitter receptors, or interfering with the multiple ion channels.